Speedy tool cut detection times in tests from 13 hours to five
Researchers from Google and the University of California Santa Cruz Genomics Institute have developed an AI-based method to analyze genome sequence data at greater speeds.
Dubbed PEPPER-Margin-DeepVariant, the method can rapidly increase disease detection times.
“Long-read sequencing has the potential to transform variant detection by reaching currently difficult-to-map regions and routinely linking together adjacent variations to enable read-based phasing,” the paper detailing the method states.
“Third-generation nanopore sequence data has demonstrated a long read length, but current interpretation methods for its novel pore-based signal have unique error profiles, making accurate analysis challenging.”
The method’s source code can be accessed on GitHub and is available under an MIT license.
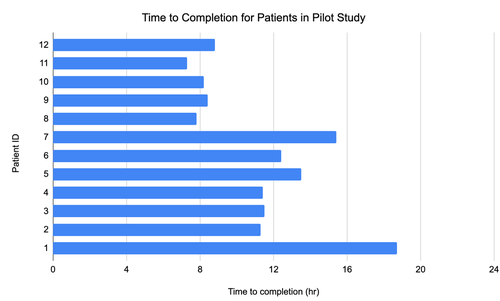
Image: Google
The method uses PEPPER, a genome inference module based on recurrent neural networks. By using the model, it enables variant calling, the process by which variants can be identified from sequence data, via DeepVariant, an analysis pipeline developed by Google.
In tests, the new method detected suspected disease-causing genomic variants in fewer than eight hours, surpassing previous records of 13 hours.
The test findings were published in the New England Journal of Medicine. Stanford University School of Medicine researchers led the tests, demonstrating the tool in a newborn intensive care unit.
Genomic data from 12 patients were analyzed, with researchers giving five diagnoses based on its breakdown. The traditional gene panel analysis takes as long as two weeks to return results.
In one instance, the researchers diagnosed a rare seizure-causing genetic disorder in a three-month-old infant in a few hours.
Following Stanford’s tests, Google’s genomics product lead and genomics software engineer lead Pi-Chuan Chang said both machine learning and algorithm development tools can “help researchers unlock more information from sequencing data.”
“We hope that our work developing and sharing these methods with those in the field of genomics will improve overall health and the understanding of biology for everyone. Working together with our collaborators, we can apply this work to real-world applications,” sates a Google blog post.
The blog notes a new partnership for Google with PacBio, a California-based biotech firm developing systems for gene sequencing.
Announced earlier in January, the partnership would see the pair collaborate on research, with PacBio to explore the use of Google’s genomic analysis, machine learning and algorithm development tools to improve its variant calls.
PacBio said it hopes to improve the utility and overall value of its HiFi data for applications like whole-genome sequencing, full-length isoform and targeted sequencing applications by integrating Google’s deep learning tech into its future product releases.
About the Author(s)
You May Also Like


.jpg?width=700&auto=webp&quality=80&disable=upscale)
.jpg?width=700&auto=webp&quality=80&disable=upscale)
.jpg?width=700&auto=webp&quality=80&disable=upscale)


.jpg?width=300&auto=webp&quality=80&disable=upscale)

.jpg?width=300&auto=webp&quality=80&disable=upscale)