DeepMind and EMBL release massive database of AI-based human protein structure predictions
Nearly the entire human proteome available for free
July 29, 2021
Nearly the entire human proteome available for free
Google's DeepMind and the European Molecular Biology Laboratory (EMBL) have created the largest ever database of human protein structures.
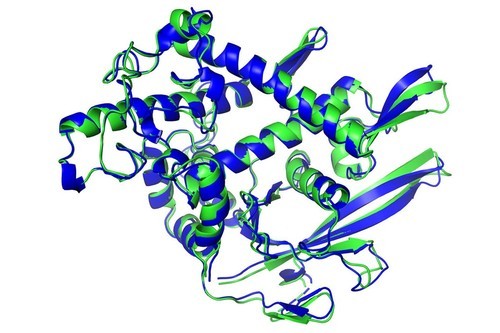
The AlphaFold database predicts the three-dimensional structures for 98.5 percent of the roughly 20,000 proteins expressed by the human genome.
Welcome to the fold
DeepMind first announced its AlphaFold algorithm in late 2020, promising a significant scientific breakthrough.
Each protein is a string made up of 20 amino acids, arranged in different orders. Their interactions with each other make the protein fold, with scientist Cyrus Levinthal estimating in 1969 that there were some 10^300 possible conformations for a typical protein.
Using conventional brute force computing to work out how to predict a protein's shape just from looking at a string of amino acids is, therefore, practically impossible.
DeepMind's AlphaFold predicts the shape of proteins after being trained on ones that were painstakingly measured by scientists over decades.
Earlier this month, the Alphabet subsidiary announced an updated version of the deep-learning neural network, claiming it was 16 times faster at generating predictions. The company also open sourced AlphaFold 2, making it free to use for the AI and healthcare community.
Now, DeepMind is following that up with a database of more than 350,000 3D protein structures, making up the 20,000 proteins expressed by the human genome.
Proteomes of 20 other organisms, including the Zebrafish, malaria parasite, and E.coli bacteria, are also in the database, with others set to be added.
"While we have presented several case studies to illustrate the type of insights that may be gained from these data, we recognize that there is still much more to uncover," the company's researchers said in a paper in Nature.
"By making our predictions available to the community we hope to enable exploration of new directions in structural bioinformatics."
Protein-folding competition CASP found that the initial version of AlphaFold was around 95 percent accurate, but the newly published paper admits there are still some proteins that are harder to predict.
"Some proportion of these will be genuine failures, where a fixed structure exists but the current version of AlphaFold does not predict it," the paper states.
"It will be crucial to develop new methods that can address the biology of these regions, for example, by predicting the structure in complex or by predicting a distribution over possible states in the cellular milieu."
The ability to predict protein structures has been heralded as a huge milestone in structural bioinformatics, which could lead to consequential advances in disease care and drug research in the decades to come.
AlphaFold is being used being tested by the Drugs for Neglected Diseases Initiative (DNDi), the Centre for Enzyme Innovation (CEI), and the University of Colorado Boulder.
About the Author(s)
You May Also Like
.jpg?width=700&auto=webp&quality=80&disable=upscale)
.jpg?width=700&auto=webp&quality=80&disable=upscale)
.jpg?width=700&auto=webp&quality=80&disable=upscale)
.jpg?width=300&auto=webp&quality=80&disable=upscale)
.jpg?width=300&auto=webp&quality=80&disable=upscale)
.jpg?width=300&auto=webp&quality=80&disable=upscale)
.jpg?width=300&auto=webp&quality=80&disable=upscale)
.jpg?width=300&auto=webp&quality=80&disable=upscale)